I spent yesterday and today working through the excellent guide by Alex on using sqlite-vss to do vector similarity search in a SQLite database.
I’m particularly interested in the benefits one can get from having these tools available locally for getting better insights into non-big datasets with a low barrier to entry.
Combining this plugin with a tool like datasette gives you a powerful data stack nearly out of the box.
Installing the sqlite-vss extension#
The ergonomics of installing and loading vector0.dylib and vss0.dylib are a little unusual.
When pip installing sqlite_vss, the extension can be loaded via
import sqlite_vss
db.enable_load_extension(True)
sqlite_vss.load(db)
However, to use it in the sqlite shell, I needed to download these dylibs to my project from the Github project releases page.
These dylibs are also downloaded with the Python library but live a bit deep in the virtualenv.
❯ ls env/lib/python3.11/site-packages/sqlite_vss
__pycache__ __init__.py vector0.dylib version.py vss0.dylib
Maybe there is a better way to automatically load these to sqlite given the installed Python packages but I haven’t come across it yet.
After downloading both of these dylibs, I could run sqlite then
.load ./vector0.dylib
.load ./vss0.dylib
then could create and query vector similarity search tables.
Running this each time I opened the sqlite shell was a chore, so I created a .sqliterc for my project
.headers on
.mode column
.load './vector0.dylib'
.load './vss0.dylib'
then loaded that each time with
sqlite3 -init .sqliterc whatever.db
I also use rlwrap to enable arrow key navigation in sqlite, described in further detail here
This is the command that does it all (data comes from the News Category Dataset used by the guide linked above)
❯ rlwrap sqlite3 -init .sqliterc database.db
-- Loading resources from .sqliterc
SQLite version 3.45.1 2024-01-30 16:01:20
Enter ".help" for usage hints.
sqlite> with matches as (
(x1...> select
(x1...> rowid,
(x1...> distance
(x1...> from vss_articles
(x1...> where vss_search(
(x2...> headline_embedding,
(x2...> (select headline_embedding from articles where rowid = 82345)
(x2...> )
(x1...> limit 5
(x1...> )
...> select
...> articles.rowid,
...> articles.headline,
...> matches.distance
...> from matches
...> left join articles on articles.rowid = matches.rowid;
rowid headline distance
------ ------------------------------------------------------ -----------------
82345 10 Books That Remind Us America Should Be For Everyone 0.0
94538 10 New Books Everyone Should Read This Summer 0.568291664123535
3490 The Best Books Of 2020 0.651552319526672
194850 50 Books That Every African American Should Read 0.654529571533203
144072 The 9 Best Books for Meaningful Change 0.721711039543152
Building the index#
I mostly followed the guide above to load the data and build the vss index. Loading the data was relatively fast as fast calculating the headline and description embeddings using the bulk script.
What took much longer (10+ minutes) was filling the vss0 virtual table. Creating the table was trivial.
create virtual table vss_articles using vss0(
headline_embedding(384),
description_embedding(384),
);
Inserting the data seemed to hang forever.
insert into vss_articles(rowid, headline_embedding, description_embedding)
select
rowid,
headline_embedding,
description_embedding
from articles;
sqlite was maxing out a core for many minutes and I had no visibility into what was happening.
It’s possible this would have completed eventually if I waited long enough, but instead, I wrote a script to build vss_articles in batches with some sqlite config changes to try and speed things up.
import os
import sqlite3
import sqlite_vss
DB = os.environ.get("DATABASE_FILE")
db = sqlite3.connect(DB)
db.enable_load_extension(True)
sqlite_vss.load(db)
BATCH_SIZE = 256
def insert_in_batches(batch_size):
db.execute("""
CREATE VIRTUAL TABLE IF NOT EXISTS vss_articles USING vss0(
headline_embedding(384),
description_embedding(384)
);
""")
cursor = db.cursor()
db.execute("PRAGMA synchronous = OFF")
db.execute("PRAGMA journal_mode = MEMORY")
db.execute("PRAGMA temp_store = MEMORY")
db.execute("PRAGMA cache_size = -64000")
# Determine total number of rows
cursor.execute("SELECT COUNT(*) FROM articles")
total_rows = cursor.fetchone()[0]
batches = (total_rows // batch_size) + (1 if total_rows % batch_size else 0)
for i in range(batches):
print(f"Batch {i+1}/{batches}")
offset = i * batch_size
db.execute("BEGIN TRANSACTION")
db.execute("""
INSERT INTO vss_articles(rowid, headline_embedding, description_embedding)
SELECT rowid, headline_embedding, description_embedding
FROM articles LIMIT ? OFFSET ?""", (batch_size, offset))
db.execute("COMMIT")
db.close()
insert_in_batches(BATCH_SIZE)
This script still took nearly 10 minutes to run, but at least I was able to see progress and have some confidence that it would eventually finish. A note (that may inform future optimizations) is that the first ~200/~800 batches loaded quite quickly. After those, the ingest pace slowed to only a few batches per second for the remainder of the run. Increasing to a batch size of 1024, got to 50% quite quickly, then slowed for the rest of the run. Overall, larger batches were clearly faster. Pushing the batch size to 2048 resulted in runtime of about 2.5 minutes. I may return to this benchmarking exercise separately.
Size of the data at various points in the process#
| Dataset | Size |
|---|---|
| Kaggle JSON | 28 MB |
| SQLite db loaded from JSON | 74.2 MB |
| SQLite with embeddings added | 860.5 MB |
| SQLite with vss virtual table | 1.83 GB |
The embeddings seem to contribute the majority of the data size. Creating the virtual table seems to be an effective doubling, which makes sense since we’re copying the embeddings to another table.
Running queries#
Finally, with the data loaded, I installed and setup a datasette instance to run queries against the vector store.
pip install datasette
datasette install datasette-sqlite-vss
I initially setup a SentenceTransformer plugin as Alex does in post and called it st_encode.
With this plugin, I started datasette with
datasette --plugins-dir=plugins database.db
I later learned I could also use the datasette-llm-embed plugin to do something similar.
Since I already had llm installed using brew for general purposes, per these instructions, in my project I ran
llm install -U llm
llm install llm-sentence-transformers
llm-sentence-transformers registers sentence-transformers/all-MiniLM-L6-v2 by default, which you can validate at
cat ~/Library/Application\ Support/io.datasette.llm/sentence-transformers.json
which (for me) outputs
[
{
"name": "all-MiniLM-L6-v2"
}
]
Or you can just run this command to confirm
❯ llm sentence-transformers register all-MiniLM-L6-v2
Error: Model all-MiniLM-L6-v2 is already registered
After restarting datasette, the following query now produces the same results as Alex’s example with the custom st_encode plugin.
with matches as (
select
rowid,
distance
from
vss_articles
where
vss_search(
headline_embedding,
llm_embed("sentence-transformers/all-MiniLM-L6-v2", "AI")
)
limit
10
)
select
articles.headline,
matches.distance
from
matches
left join articles on articles.rowid = matches.rowid
You can navigate to http://127.0.0.1:8001/database to try it yourself.
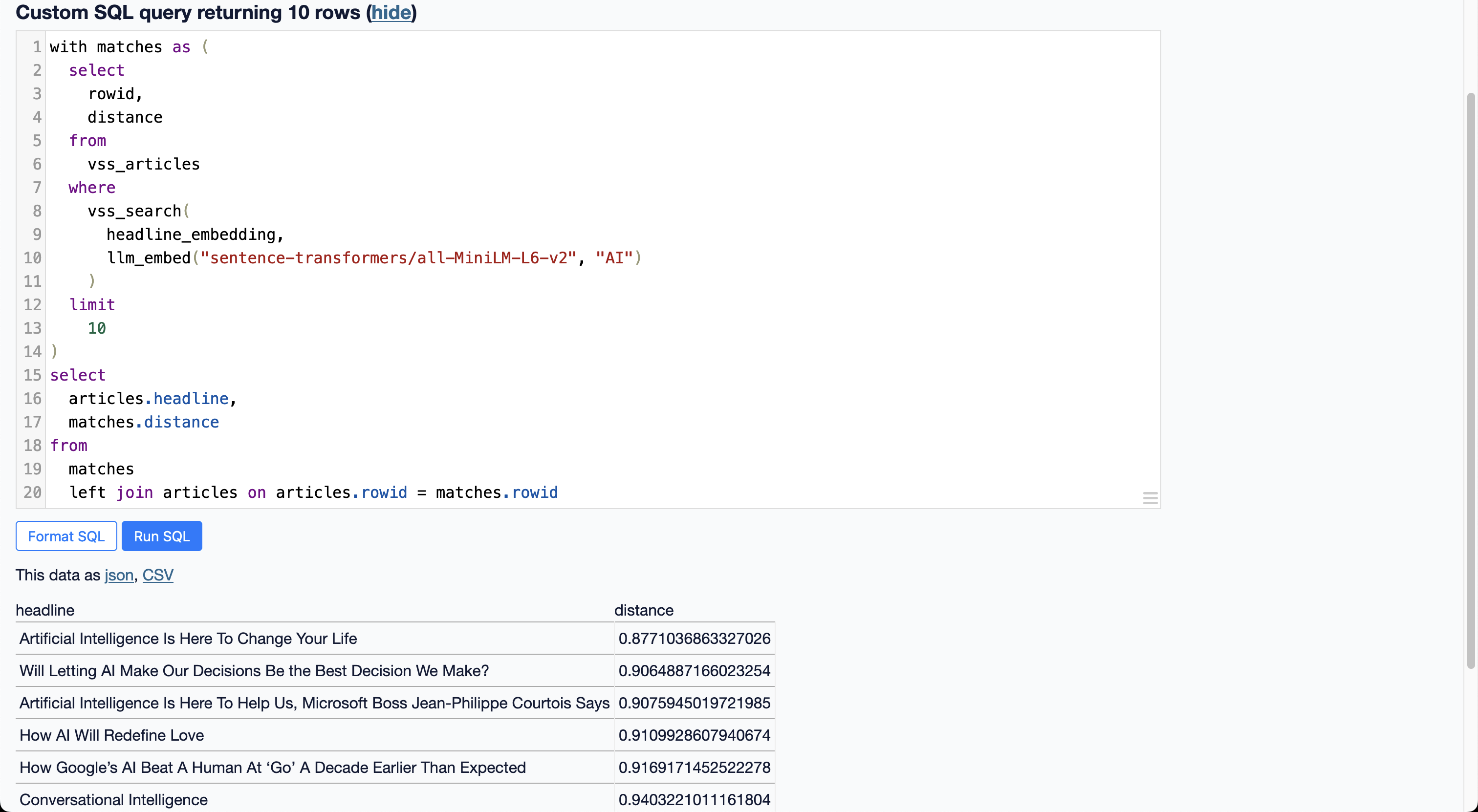
It works pretty well! We find several article titles that relate to “AI” even though the exact string “AI” doesn’t appear in all of them.